You can connect with me on LinkedIn to discuss collaborations and work opportunities.
The problem
We want to find out which are the top #5 American airports with the largest average (mean) delay on domestic flights.

Data
We will be using the Data Expo 2009: Airline on time data dataset from the Harvard Dataverse. The data consists of flight arrival and departure details for all commercial flights within the USA, from October 1987 to April 2008. This is around 120 million records, divided in 22 CSV files, one per year, and 4 auxiliary CSV files that we will not use here. The total size on disk of the dataset is around 13 Gb. The original data comes compressed, but the decompression part is not considered part of the pipeline here.
Environment
The available hardware to do the job are a single computer with the next specs:
- Intel(R) Core(TM) i7-8550U CPU @ 1.80GHz
- Memory: LPDDR3 15820512 kB (16 Gb) 2133 MT/s, no swap
- Disk: KXG50ZNV512G NVMe TOSHIBA 512GB (ext4 non-encrypted partition)
- OS: Linux 5.19.9 (Arch distribution, only KDE Plasma and a single Konsole session running, using 610 Mb of RAM)
The versions of the software used in the post are:
Naive approach
A first try at solving the problem, could involve using pandas and loading the data with the next code:
import pandas
df = pandas.concat((pandas.read_csv(f'{year}.csv') for year in range(1987, 2009)))
Unfortunately, this is likely to raise a MemoryError (or restart the kernel if you are
using Jupyter), unless you have a huge amount of RAM in your system.
The rest of this article describes different ways to avoid this error, and how to make your
code faster and more efficient with simple options.
Pure python
In this particular case, we do not really need to load all the data into memory to get the average delay for each airport. We can discard the rows as we read them, just keeping track of the cumulative delay and the number of rows read for each airport. So, once all rows have been processed, we can simply compute the mean by dividing the total delay by the number of flights.
This could be an implementation:
import csv
import datetime
import heapq
import operator
USE_COLS = 'Origin', 'Year', 'Month', 'DayofMonth', 'CRSDepTime', 'DepTime'
airports = {}
for year in rage(1987, 2009):
with open(f'../data/{year}.csv', errors='ignore') as f:
reader = csv.reader(f)
header = {name: position
for position, name
in enumerate(next(reader))
if name in USE_COLS}
for row in reader:
if row[header['CRSDepTime']] == 'NA' or row[header['DepTime']] == 'NA':
continue
year, month, day = (int(row[header['Year']]),
int(row[header['Month']]),
int(row[header['DayofMonth']]))
try:
scheduled_dep = datetime.datetime(year, month, day,
int(row[header['CRSDepTime']][:-2] or '0'),
int(row[header['CRSDepTime']][-2:]))
actual_dep = datetime.datetime(year, month, day,
int(row[header['DepTime']][:-2] or '0'),
int(row[header['DepTime']][-2:]))
except ValueError:
continue
delay = (actual_dep - scheduled_dep).total_seconds() / 3600.
if delay < -2.:
delay = 24. - delay
if row[header['Origin']] not in airports:
airports[row[header['Origin']]] = [1, delay]
else:
airports[row[header['Origin']]][0] += 1
airports[row[header['Origin']]][1] += delay
print(dict(heapq.nlargest(5,
((airport, total_delay / count) for airport, (count, total_delay) in airports.items()),
operator.itemgetter(1))))
Note that if we were interested in the median instead of the mean, it would require a more complex approach, and we would not be able to discard the individual delays immediately.
With this method we solve the memory problem, as we just keep a very limited amount of information in RAM at a time. Running this code will not take much more than 1 Mb of heap memory.
This code takes around 7 minutes to complete.
PyPy
CPython (the Python interpreter used when we type python <our-program>) is very fast at
what it does, but what it does it involves a lot of magic to allow our code to be more
readable, and let programmers forget about many internal details.
PyPy is a different interpreter for Python code, that takes a different approach than CPython, using a just in time compiler, and for some tasks it can do the same job, but faster.
Running the code above in PyPy will consume much more memory, around 40 Mb (which for modern standards is still almost nothing), and will run in approximately 4 minutes and 40 seconds. This is two-thirds of the time, compared to CPython.
pandas
Back to the pandas world, there are few things we can do to save memory and time when our data is big, and we need to take performance into consideration.
Some things that helped in this particular case:
- Load only the columns that you are going to use
- Specify the types of the columns you are reading, and use the most efficient for your data
- Make sure you never make the Python interpreter run loops. This means writing your own loops,
as well as telling pandas to run loops with the interpreter (e.g.
DataFrame.apply) - Avoid copying the data (e.g.
DataFrame.copy) - Filter the data as soon as possible, and don't keep references to unused data structures
(
DataFrame,Series, etc) if they are big
This is one possible implementation that will solve the MemoryError problem,
following the above guidelines:
import pandas
LOAD_COLS = 'Origin', 'Year', 'Month', 'DayofMonth', 'CRSDepTime', 'DepTime'
df = pandas.concat((pandas.read_csv(f'../data/{fname}.csv',
usecols=LOAD_COLS,
encoding_errors='ignore',
dtype={'Origin': 'category',
'Year': 'uint16',
'Month': 'uint8',
'DayofMonth': 'uint8',
'CRSDepTime': 'uint16',
'DepTime': 'UInt16'})
for fname
in range(1987, 2009)),
ignore_index=True)
date = pandas.to_datetime(df[['Year', 'Month', 'DayofMonth']].rename(columns={'DayofMonth': 'Day'}))
df['scheduled_dep'] = date + pandas.to_timedelta((df['CRSDepTime'] // 100) * 60 + (df['CRSDepTime'] % 100),
unit='minutes')
df['actual_dep'] = date + pandas.to_timedelta((df['DepTime'] // 100) * 60 + (df['DepTime'] % 100),
unit='minutes')
del date
df = df[['Origin', 'scheduled_dep', 'actual_dep']]
df['delay'] = (df['actual_dep'] - df['scheduled_dep']).dt.total_seconds() / 60 / 60
df['delay'] = df['delay'].where(df['delay'] > - 2, 24 - df['delay'])
print(df.groupby('Origin')['delay'].mean().sort_values(ascending=False).head(5))
This program completes in around 2 minutes and 45 seconds. This is around 60% of what PyPy
took. In this case, pandas will load all data into memory, and it will require around 8.1 Gb
of memory in its peak (the peak happens during pandas.concat).
In this case, the pipeline can be divided in two different parts. The part where we load the data into memory, and the part where we transform the data as needed and extract the information we want to know. It is worth analyzing how long it takes for each part to run, so we do not spend too much time optimizing code that will not have a big impact in the total time.
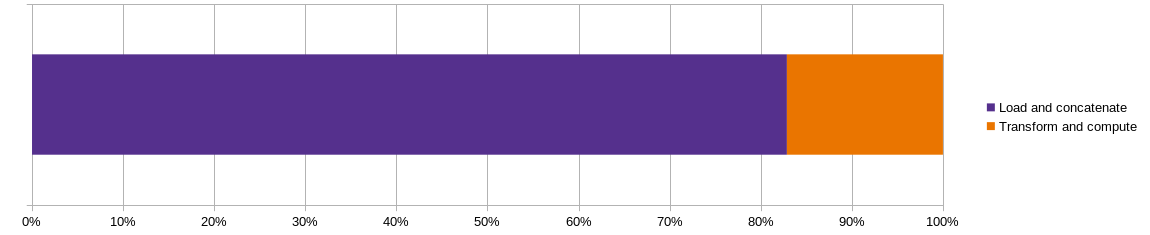
We can see that more than 80% of the time our pipeline is taking to complete, is spent loading the data from the CSV files, and combining the CSV files into a single dataframe. This is the part we will be focusing in the next sections.
pandas with PyArrow
pandas supports different engines when loading data from CSV files (and also other formats).
This means that in practice pandas has three different pandas.read_csv functions, but
wraps them in a single one with a parameter to decide which one needs to run. You can think
about read_csv like if this was implemented:
def read_csv(fname, engine='c'):
if engine == 'c':
return read_csv_with_pandas_implementation_in_c(fname)
elif engine == 'python':
return read_csv_with_pandas_implementation_in_python(fname)
elif engine == 'pyarrow':
return pyarrow.read_csv(fname).to_pandas()
else:
raise ValueError
The python engine for read_csv is slower and should be avoided in most cases. But the
PyArrow implementation can perform better, and it just requires to install PyArrow
(e.g. mamba install pyarrow), and call read_csv with engine='pyarrow'.
With the PyArrow engine, our code above runs in around 1 minute and 10 seconds. This is
less than half than the original code, around 42%. The memory consumption at the peak is
the same, since it happens at pandas.concat, which will receive the same dataframes
(even if they were generated with different implementations, the resulting dataframes
will be the same).
The PyArrow implementation uses multithreading, and is able to make use of CPUs with more than one core when loading the data. The speed up is clearly not 8 times faster (as my laptop has 8 cores), since the bottleneck is likely to be reading for the disk.
NVMe disks like the one used here are SSD disks much faster than mechanical disks. I guess using multithreading in mechanical disks may not make code faster, and other techniques like keeping data compressed in disk, and only decompress directly in memory as part of the pipeline can be tried. This would reduce the amount of bytes read from disk, and it may be the case that the time to decompress the data is lower than the time saved by reading less data. But that would depend on every environment.
PyArrow
There is something else we can still do, which is using PyArrow directly, instead of using it from pandas. The code to perform the operations once the data is loaded would be the same, but the loading of the dataset would look like this:
import pyarrow
import pyarrow.csv
COLUMN_TYPES = {'Origin': pyarrow.dictionary(pyarrow.int32(),
pyarrow.string()),
'Year': pyarrow.uint16(),
'Month': pyarrow.uint8(),
'DayofMonth': pyarrow.uint8(),
'CRSDepTime': pyarrow.uint16(),
'DepTime': pyarrow.uint16()}
tables = []
for year in range(1987, 2009):
tables.append(pyarrow.csv.read_csv(
f'../data/{year}.csv',
convert_options=pyarrow.csv.ConvertOptions(
include_columns=COLUMN_TYPES,
column_types=COLUMN_TYPES)))
df = pyarrow.concat_tables(tables).to_pandas()
This solution will take around 50 seconds, around 70% of the time taken by the pandas with PyArrow engine implementation. And the memory required at the peak is 7.5 Gb (93% of the required by the pandas/PyArrow implementation).
The time to read CSV files should be around the same whether we do it in PyArrow, or in
pandas with the PyArrow engine. There will be an overhead in pandas, but should be negligible.
The main different with both implementations will be when concatenating the dataframes. The
PyArrow implementation is concatenating Arrow tables (or record batches), which is a more
optimized and faster operation than pandas.concat, which in general will concatenate
NumPy arrays.
PyArrow by year
The previous approach seems to start looking good enough, at least if you have the 7.5 Gb of memory available. But can we do better? We saw in the Python and PyPy approaches at the beginning, did not need to have all the data loaded in memory, for this particular example of computing the mean. We are loading it because pandas makes our code significantly faster, than iterating over the data in Python, which is costly. But there is a hybrid solution we can do, which is to load only the data for a single year at a time.
So, the new approach will be mostly the same as before, but for each year, except that in the last operation of computing the mean, we are going to save the cumulative delay and the number of flights for each airport. And after processing all the years, we will take the result of every year, add them up, and compute the mean delays by dividing the total delay by the number of flights for each airport.
The code is slightly more complex than before, but still reasonably simple:
import functools
import pyarrow
import pyarrow.csv
import pandas
COLUMN_TYPES = {'Origin': pyarrow.dictionary(pyarrow.int32(),
pyarrow.string()),
'Year': pyarrow.uint16(),
'Month': pyarrow.uint8(),
'DayofMonth': pyarrow.uint8(),
'CRSDepTime': pyarrow.uint16(),
'DepTime': pyarrow.uint16()}
results = []
for year in range(1987, 2009):
df = pyarrow.csv.read_csv(
f'../data/{year}.csv',
convert_options=pyarrow.csv.ConvertOptions(
include_columns=COLUMN_TYPES,
column_types=COLUMN_TYPES)).to_pandas()
date = pandas.to_datetime(df[['Year', 'Month', 'DayofMonth']].rename(columns={'DayofMonth': 'Day'}))
df['scheduled_dep'] = date + pandas.to_timedelta((df['CRSDepTime'] // 100) * 60 + (df['CRSDepTime'] % 100),
unit='minutes')
df['actual_dep'] = date + pandas.to_timedelta((df['DepTime'] // 100) * 60 + (df['DepTime'] % 100),
unit='minutes')
df = df[['Origin', 'scheduled_dep', 'actual_dep']]
df['delay'] = (df['actual_dep'] - df['scheduled_dep']).dt.total_seconds() / 60 / 60
df['delay'] = df['delay'].where(df['delay'] > - 2, 24 - df['delay'])
results.append(df.groupby('Origin')['delay'].agg(['sum', 'count']))
df = functools.reduce(lambda x, y: x.add(y, fill_value=0), results)
df['mean'] = df['sum'] / df['count']
print(df['mean'].sort_values(ascending=False).head(5))
This solution is faster, it takes around 37 seconds (74% of the time it took loading all the data at once). The time to read the files should stay the same, and the time to compute the operations should be slightly slower, since there are synergies in operating with all data at once. But the main difference should be in the concatenation that we do not perform anymore. That operation requires allocating memory and moving data, which are slow operations.
As expected, the memory consumption at the peak is lower, as we do not need to keep all the data loaded at once. The peak memory allocated in the heap is around 900 Mb, this is around 12% of what it consumed when all the data was loaded at once. This of course will depend on the number of files in which the data is split.
pandas by year with multiprocessing
For the last example, I will be back to using pandas again, with its native implementation
of read_csv (the c engine). We have seen how PyArrow has been faster, and one of the
main reasons is that PyArrow is using all the available CPUs in the machine, while the
pandas implementation uses only one core. In our last example, we saw that we can process
just one CSV file at a time, and that makes things not only more memory efficient, but
also faster. We will use the same approach, but will process the CSV files in parallel.
This is mostly for illustration, and I would not in general recommend this approach for
this problem. We will use the Python multiprocessing module, and a Pool with as many
workers as CPUs in the machine (8 in my case). Other than that, the algorithm is the same
as in the previous example.
import functools
import multiprocessing
import pandas
LOAD_COLS = 'Origin', 'Year', 'Month', 'DayofMonth', 'CRSDepTime', 'DepTime'
def read_one_csv(year):
df = pandas.read_csv(f'../data/{year}.csv',
engine='c',
usecols=LOAD_COLS,
encoding_errors='ignore',
dtype={'Origin': 'category',
'Year': 'uint16',
'Month': 'uint8',
'DayofMonth': 'uint8',
'CRSDepTime': 'uint16',
'DepTime': 'UInt16'})
date = pandas.to_datetime(df[['Year', 'Month', 'DayofMonth']].rename(columns={'DayofMonth': 'Day'}))
df['scheduled_dep'] = date + pandas.to_timedelta((df['CRSDepTime'] // 100) * 60 + (df['CRSDepTime'] % 100),
unit='minutes')
df['actual_dep'] = date + pandas.to_timedelta((df['DepTime'] // 100) * 60 + (df['DepTime'] % 100),
unit='minutes')
df = df[['Origin', 'scheduled_dep', 'actual_dep']]
df['delay'] = (df['actual_dep'] - df['scheduled_dep']).dt.total_seconds() / 60 / 60
df['delay'] = df['delay'].where(df['delay'] > - 2, 24 - df['delay'])
return df.groupby('Origin')['delay'].agg(['sum', 'count'])
pool = multiprocessing.Pool()
df = functools.reduce(lambda x, y: x.add(y, fill_value=0),
pool.map(read_one_csv, range(1987, 2009)))
df['mean'] = df['sum'] / df['count']
print(df['mean'].sort_values(ascending=False).head(5))
This code runs in 53 seconds, this is like one-third of the original pandas code with
the c engine, but significantly slower than the equivalent implementation with PyArrow.
If the CPU was the bottleneck, we would expect to have the code running around 8 times
faster (assuming CSV files have the same size, and the overhead for multiprocessing is
not significant). But as expected, the disk may be the bottleneck, and more CPUs are not
helping much after the third (with 3 processes, the time increases from 53 seconds to 66).
The memory usage is more complex to analyze in this case. Every process is using its own space in the memory heap. There is not a single peak in memory, but one for each process, which may or may not happen at the same time, depending on different factors. Each process requires around 400 Mb of memory at its peak. In the best scenario that will be the amount of memory we require for the pipeline. In the worst all the peaks happen at the same time, and we will have 8 times that value. In the chart below, I use 4 times the peak process, but the number is probably meaningless without understanding the details.
Summary
We have seen how one problem that initially could not be solved for lack of memory, could finally be solved in a reasonable time and with reasonable memory requirements.
The code we used has always been in Python, and in all the cases it has been short and not
too complex. We saw how in some cases we get big gains with no changes to the code (e.g.
using PyPy) or with minimal changes (e.g. using the PyArrow read_csv engine instead of
the default c one). In some other cases the changes were bigger, but we could still
get much better performance without fundamental changes.
All the approaches covered here are just a minimal part of the options available to make our code run faster and with less memory. I considered only options based on Python and pandas. But there are many other options that could be explored:
- Using other programming languages, such as R, Julia, Rust or C++
- Using pandas alternatives such as Vaex or Polars
- Using different formats, such as Parquet, HDF or ORC
- Using a cluster instead of a single computer, and use Dask, Modin or Koalas
- Use Python with other just in time compilers, such as Bodo or Numba
And probably many other reasonable options. I may write about some of these options in future blog posts.
Finally, here you have couple of charts with the summary of the results:
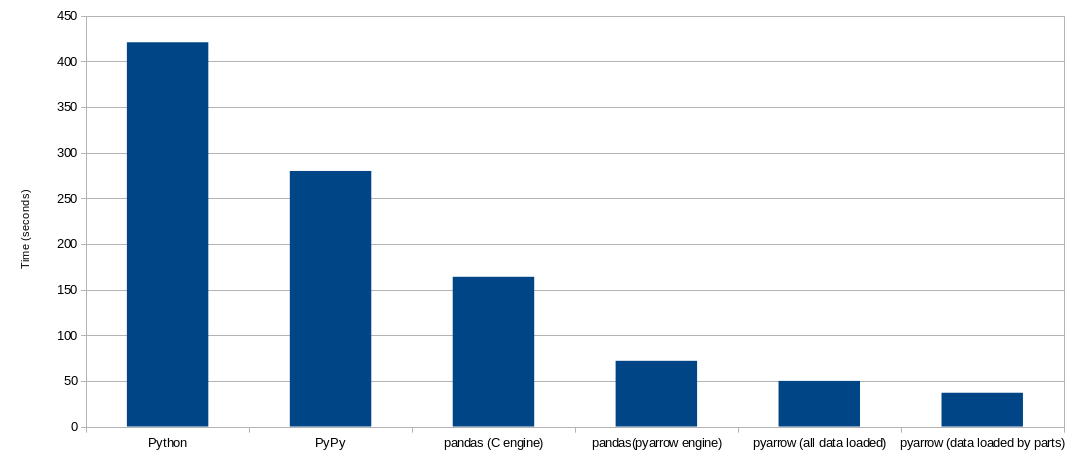
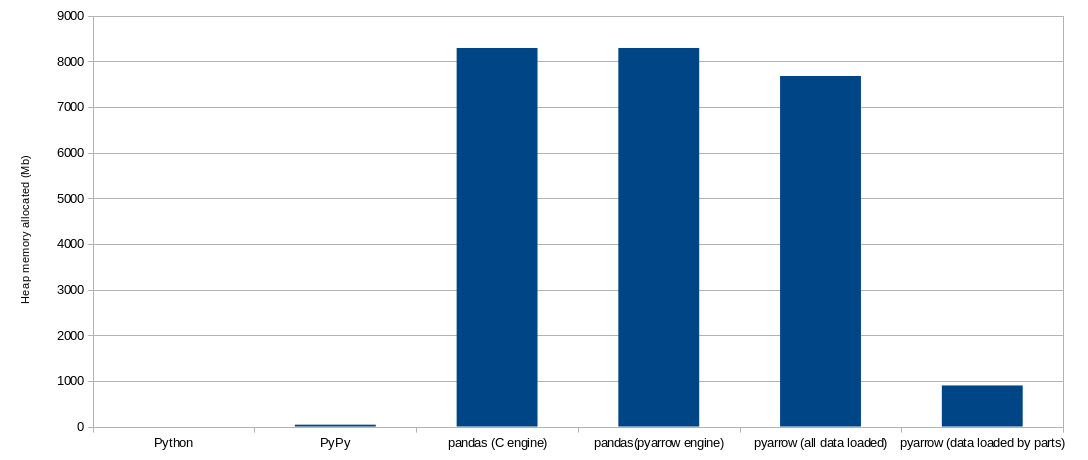
Further reading
If you fly from Muskegon County Airport (MKG), in Michigan, I recommend you to bring a good book, since waiting 6 hours for your flight does not seem unlikely. ;) If you enjoyed this post, I recommend High Performance Python. It does not only provide a lot of useful information on how to make your Python code more efficient, but it is also very enjoyable to read.
You can also be interested in my Demystifying pandas internals talk, at PyData London 2018. It provides more context on the Python vs pandas part.
You can also follow my datapythonista Telegram channel. I post there when I write blog posts, including this one and the follow ups. I also post there shorter technical notes not published elsewhere, and a good amount of software and data memes. :)